Operations with Frovedis DataFrame
Nov 1, 2021, Shoichiro Yokotani, Application Development Expert, AI Platform division
By using Python, you can select the data format such as List, Dictionary, Tuple in the standard library as the data destination. In addition, the Python libraries pandas and Numpy provide flexible operation functions.
This article will focus specifically on pandas. pandas provides data analysts with functions for formating labeled data structures.
After merging, aggregating, and slicing multiple tables using pandas, you can check the statistical information of your data and perform analysis using data analysis algorithms. In order to handle data stored in List or Dictionary format, it is necessary to create processing code. With pandas, various processes can be realized using the functions of DataFrame. It is faster to use the pandas method than to make changes to the List or Dictionary format data by loop processing.
Also, the larger the data, the greater the difference in processing speed.
When handling relatively small data on SX-Aurora TSUBASA, you can execute analysis on the Vector Engine by getting the data with pandas DataFrame and converting it to Frovedis DataFrame. At this time, the data is transferred from the x86 CPU main memory to the Vector Engine memory.
The functionality provided by the Frovedis DataFrame is equivalent to a subset of the pandas version. You can use functions for exchanging data with pandas DataFrame as well as basic operation functions such as Select, Join, Sort, and Groupby. Data shaping can be made flexible by linking with mathematical arithmetic processing using Numpy's multidimensional array object, handling of time series data of pandas, and data input / output function.
Let's take a look at some examples of working with Frovedis DataFrame while using Jupyter notebook. First, perform data operations such as select, sort, and group by with small data.
Frovedis DataFrame operation example using sample data
In [1]:
|
import os import numpy as np import pandas as pd from frovedis.exrpc.server import FrovedisServer from frovedis.dataframe.df import FrovedisDataframe |
Starting Frovedis server
In [2]:
|
FrovedisServer.initialize("mpirun -np 2 {}".format(os.environ['FROVEDIS_SERVER'])) |
Out[2]:
'[ID: 1] FrovedisServer (Hostname: handson02, Port: 42383) has been initialized with 2 MPI processes.'
Two types of sample data are prepared in dictionary format. Converting to Frovedis DataFrame via pandas DataFrame. Data is transferred to the Vector Engine's memory as it is converted from pandas to Frovedis DataFrame.
In [3]:
|
peopleDF ={ 'Ename': ['Michael', 'Andy', 'Tanaka', 'Raul', 'Yuta'], 'Age': [29, 30, 27, 19, 31], 'Country': ['USA', 'England', 'Japan', 'France', 'Japan'], } |
In [4]:
|
countryDF ={ 'Ccode': [1, 2, 3, 4], 'Country': ['USA', 'England', 'Japan', 'France'], } |
In [5]:
|
pdf1 = pd.DataFrame(peopleDF) pdf2 = pd.DataFrame(countryDF) fdf1 = FrovedisDataframe(pdf1) fdf2 = FrovedisDataframe(pdf2) |
Showing Frovedis Data Frame.
In [6]:
|
# display created frovedis dataframes print ("* print Frovedis DataFrame") print (fdf1.to_pandas_dataframe()) print (fdf2.to_pandas_dataframe()) |
* print Frovedis DataFrame
| Ename | Age | Country | |
| Index | |||
| 0 | Michael | 29 | USA |
| 1 | Andy | 30 | England |
| 2 | Tanaka | 27 | Japan |
| 3 | Raul | 19 | France |
| 4 | Yuta | 31 | Japan |
| Ename | Country | |
| Index | ||
| 0 | 1 | USA |
| 1 | 2 | England |
| 2 | 3 | Japan |
| 3 | 4 | France |
Column is selected by specifying the column name.
In [7]:
|
# select demo print ("* select Ename and Age") print (fdf1[["Ename","Age"]].to_pandas_dataframe()) |
* select Ename and Age
| Ename | Age | |
| 0 | Michael | 29 |
| 1 | Andy | 30 |
| 2 | Tanaka | 27 |
| 3 | Raul | 19 |
| 4 | Yuta | 31 |
Filtering by specifying "age" and "country name".
In [8]:
|
# filter demo print ("* filter by Age > 19 and Contry == 'Japan'") print (fdf1[fdf1.Age > 19 and fd1.Country == 'Japan'].to_pandas_dataframe()) |
* filter by Age > 19 and Country == 'Japan'
| Ename | Age | Country | |
| 0 | Tanaka | 27 | Japan |
| 1 | Yuta | 31 | Japan |
Sorting by "age".
In [9]:
|
# sort demo print ("* sort by Age (descending order)") print (fdf1.sort("Age",ascending = False).to_pandas_dataframe()) # single column, descending |
* sort by Age (descending order)
| Ename | Age | Country | |
| 0 | Yuta | 31 | Japan |
| 1 | Andy | 30 | England |
| 2 | Michael | 29 | USA |
| 3 | Tanaka | 27 | Japan |
| 4 | Raul | 19 | France |
Sorting by specifying multiple columns.
In [10]:
|
print ("* sort by Country and Age") print (fdf1.sort(["Country", "Age"]).to_pandas_dataframe()) # multiple column |
* sort by Country and Age
| Ename | Age | Country | |
| 0 | Andy | 30 | England |
| 1 | Raul | 19 | France |
| 2 | Tanaka | 27 | Japan |
| 3 | Yuta | 31 | Japan |
| 4 | Michael | 29 | USA |
Grouping by "country name", and totaling the maximum, minimum, and average ages in each group.
In [11]:
|
# groupby demo print ("* groupby Country and max/min/mean of Age and count of Ename") out = fdf1.groupby('Country') out = out.agg({'Age': ['max','min','mean']}) out2 = out[["Country","max_Age","min_Age","mean_Age"]] print(out2.to_pandas_dataframe()) |
* groupby Country and max/min/mean of Age and count of Ename
| Country | max_Age | min_Age | mean_Age | |
| 0 | England | 30 | 30 | 30.0 |
| 1 | Japan | 31 | 27 | 29.0 |
| 2 | France | 19 | 19 | 19.0 |
| 3 | USA | 29 | 29 | 29.0 |
Renaming the column.
In [12]:
|
# renaming demo print ("* rename Country to Cname") fdf3 = fdf2.rename({'Country' : 'Cname'}) print(fdf3.to_pandas_dataframe()) |
* rename Country to Cname
| Ccode | Cname | |
| index | ||
| 0 | 1 | USA |
| 1 | 2 | England |
| 2 | 3 | Japan |
| 3 | 4 | France |
Two DataFrames are integrated by specifying the column name.
In [13]:
|
# join after column renaming print ("* merge (join) two tables") out = fdf1.merge(fdf3, left_on="Country", right_on="Cname") # with defaults print(out.to_pandas_dataframe()) |
* merge (join) two tables
| Ename | Age | Country | Ccode | Cname | |
| index | |||||
| 0 | Michael | 29 | USA | 1 | USA |
| 1 | Andy | 30 | England | 2 | England |
| 2 | Tanaka | 27 | Japan | 3 | Japan |
| 3 | Raul | 19 | France | 4 | France |
| 4 | Yuta | 31 | Japan | 3 | Japan |
Integration and sorting of two DataFrames. Displaying the selected result.
In [14]:
|
# operation chaining: join -> sort -> select -> show print ("* chaining: merge two tables, sort by Age, and select Age, Ename and Country") out = fdf1.merge(fdf3, left_on="Country", right_on="Cname") \ .sort("Age")[["Age", "Ename", "Country"]] print(out.to_pandas_dataframe()) |
* chaining: merge two tables, sort by Age, and select Age, Ename and Country
| Age | Ename | Country | |
| 0 | 19 | Raul | France |
| 1 | 27 | Tanaka | Japan |
| 2 | 29 | Michael | USA |
| 3 | 30 | Andy | England |
| 4 | 31 | Yuta | Japan |
Displaying statistical information of data in DataFrame.
In [15]:
|
# column statistics print ("describe: ") print (fdf1.describe()) print ("\n") |
describe:
| Age | |
| count | 5.000000 |
| mean | 27.200000 |
| std | 4.816638 |
| sum | 136.000000 |
| min | 19.000000 |
| max | 31.000000 |
Creating a new Frovedis DataFrame "joined" by integrating pandas DataFrame and Frovedis DataFrame.
In [16]:
|
# merging with panda dataframe print ("* merge with pandas table") pdf2.rename(columns={'Country' : 'Cname'},inplace=True) joined = fdf1.merge(pdf2, left_on='Country', right_on='Cname') print(joined.to_pandas_dataframe()) |
* merge with pandas table
| Ename | Age | Country | Ccode | Cname | |
| Index | |||||
| 0 | Michael | 29 | USA | 1 | USA |
| 1 | Andy | 30 | England | 2 | England |
| 2 | Tanaka | 27 | Japan | 3 | Japan |
| 3 | Raul | 19 | France | 4 | France |
| 4 | Yuta | 31 | Japan | 3 | Japan |
Converting Frovedis DataFrame to pandas DataFrame.
In [17]:
|
# conversion print ("* convert Frovedis DataFrame to Pandas DataFrame") print (df1.to_pandas_dataframe()) print ("\n") |
* convert Frovedis DataFrame to Pandas DataFrame
| Ename | Age | Country | |
| Index | |||
| 0 | Michael | 29 | USA |
| 1 | Andy | 30 | England |
| 2 | Tanaka | 27 | Japan |
| 3 | Raul | 19 | France |
| 4 | Yuta | 31 | Japan |
In [18]:
|
# conversion FrovedisServer.shut_down() |
Next, let's look at an operation example using Kaggle's Covid-19 vaccine data.
Processing with the Kaggle COVID-19 World Vaccination Progress dataset
Frovedis DataFrame Operation Example using Covid-19 Vaccine Data Set
in [1]:
|
import pandas as pd import numpy as np from datetime import datetime import time, os import itertools import matplotlib.pyplot as plt from frovedis.exrpc.server import FrovedisServer from frovedis.dataframe.df import FrovedisDataframe as fd |
Starting Frovedis Server.
In [2]:
|
FrovedisServer.initialize("mpirun -np 2 {}".format(os.environ['FROVEDIS_SERVER'])) |
Out[2]:
'[ID: 1] FrovedisServer (Hostname: handson02, Port: 39739) has been initialized with 2 MPI processes.'
Loading Vaccine Data Set into pandas DataFrame.
In [3]:
| manufacturers = pd.read_table('../data/country_vaccinations_by_manufacturer.csv', sep=',', engine='python').dropna() |
In [4]:
| manufacturers.head() |
Out[4]:
| location | date | vaccine | total_vaccinations | |
| 0 | Chile | 2020-12-24 | Pfizer/BioNTech | 420 |
| 1 | Chile | 2020-12-25 | Pfizer/BioNTech | 5198 |
| 2 2 | Chile | 2020-12-26 | Pfizer/BioNTech | 8338 |
| 3 | Chile | 2020-12-27 | Pfizer/BioNTech | 8649 |
| 4 | Chile | 2020-12-28 | Pfizer/BioNTech | 8649 |
Converting from pandas to Frovedis DataFrame. At this time, data is transferred to the memory of the vector engine.
In [5]:
| fd_manufacturers = fd(manufacturers) |
Inoculating countries included in the Frovedis Data Frame are being extracted. After that, the extracted data is converted to NumPy, converted to a list, and displayed.
In [6]:
|
location = fd_manufacturers['location') location = location.to_pandas_dataframe().to_numpy() x = set(list(itertools.chain.from_iterable(location))) print(x) |
Displaying Frovedis DataFrame stats.
In [7]:
| fd_manufacturers.describe() |
Out[7]:
| total_vaccinations | |
| count | 3.491000e+03 |
| mean | 4.885988e+06 |
| std 2 | 1.635967e+07 |
| sum | 1.705698e+10 |
| min | 0.000000e+00 |
| max | 1.341169e+08 |
Getting Data by country from Frovedis DataFrame.
In [8]:
|
fd_US = fd_manufacturers[fd_manufacturers.location=='United States'] fd_Germany = fd_manufacturers[fd_manufacturers.location=='Germany'] fd_Chile = fd_manufacturers[fd_manufacturers.location=='Chile'] fd_Chile.to_pandas_dataframe().head() |
| location | date | vaccine | total_vaccinations | |
| 0 | Chile | 2020-12-24 | Pfizer/BioNTech | 420 |
| 1 | Chile | 2020-12-25 | Pfizer/BioNTech | 5198 |
| 2 2 | Chile | 2020-12-26 | Pfizer/BioNTech | 8338 |
| 3 | Chile | 2020-12-27 | Pfizer/BioNTech | 8649 |
| 4 | Chile | 2020-12-28 | Pfizer/BioNTech | 8649 |
The total number of vaccinations is being calculated for each type of vaccine. Using Frovedis groupby and agg, the total is calculated and displayed for each vaccine. Here, the aggregated results of U.S., Germany, and Chile are displayed.
In [9]:
|
fdUS_out = fd_US.groupby('vaccine'.agg({'total_vaccinations': ['sum']}) fdGermany_out = fd_Germany.groupby('vaccine'.agg({'total_vaccinations': ['sum']}) fdChile_out = fd_Chile.groupby('vaccine'.agg({'total_vaccinations': ['sum']}) |
In [10]:
|
fdUS_out.sort("sum_total_vaccinations",ascending=False).to_pandas_dataframe().head() |
| vaccine | sum_total_vaccinations | |
| 0 | Pfizer/BioNTech | 6572931045 |
| 1 | Moderna | 5789377678 |
| 2 | Johnson&Johnson | 298233690 |
In [11]:
|
fdGermany_out.sort("sum_total_vaccinations",ascending=False).to_pandas_dataframe().head() |
| vaccine | sum_total_vaccinations | |
| 0 | Pfizer/BioNTech | 1006759304 |
| 1 | Oxford/AstraZeneca | 227902088 |
| 2 | Moderna | 63056380 |
| 3 | Johnson&Johnson | 68781 |
In [12]:
|
fdChile_out.sort("sum_total_vaccinations",ascending=False).to_pandas_dataframe().head() |
| vaccine | sum_total_vaccinations | |
| 0 | Sinovac | 656309318 |
| 1 | Pfizer/BioNTech | 82842894 |
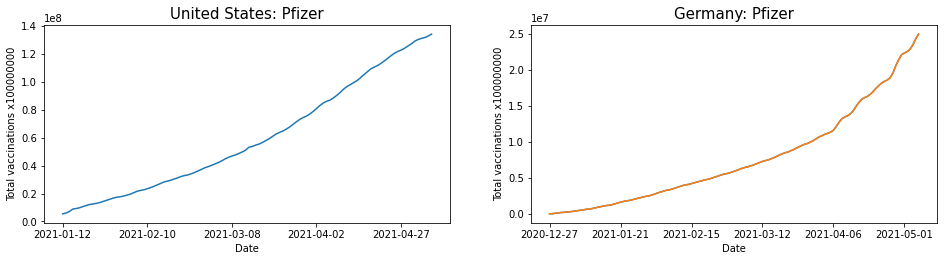
Extracting the number of vaccinations made by Pfizer for U.S. and Germany.
In [13]:
|
US_Pfizer = fd_US[fd_US.vaccine=='Pfizer/BioNTech'] US_Pfizer.to_pandas_dataframe().head() |
| location | date | vaccine | total_vaccinations | |
| 0 | United States | 2021-01-12 | Pfizer/BioNTech | 5488697 |
| 1 | United States | 2021-01-13 | Pfizer/BioNTech | 6025872 |
| 2 | United States | 2021-01-15 | Pfizer/BioNTech | 7153268 |
| 3 | United States | 2021-01-19 | Pfizer/BioNTech | 8874811 |
| 4 | United States | 2021-01-20 | Pfizer/BioNTech | 9281063 |
In [14]:
|
Germany_Pfizer = fd_Germany[fd_Germany.vaccine=='Pfizer/BioNTech'] Germany_Pfizer.to_pandas_dataframe().head() |
| location | date | vaccine | total_vaccinations | |
| 0 | Germany | 2020-12-27 | Pfizer/BioNTech | 24473 |
| 1 | Germany | 2020-12-28 | Pfizer/BioNTech | 42813 |
| 2 | Germany | 2020-12-29 | Pfizer/BioNTech | 92363 |
| 3 | Germany | 2020-12-30 | Pfizer/BioNTech | 154903 |
| 4 | Germany | 2020-12-31 | Pfizer/BioNTech | 204951 |
Converting time series data of Pfizer vaccine inoculation in U.S. and Germany to Pandas DataFrame. The graph of the converted data is shown below.
In [15]:
|
US_data = US_Pfizer.to_pandas_dataframe() Germany_data = Germany_Pfizer.to_pandas_dataframe() fig = plt.figure(figsize=(16,8)) ax1 = fig.add_subplot(2,2,1) ax2 = fig.add_subplot(2,2,2) x1 = range(0,len(US_data['date']),25) ax1.plot(US_data['date']),US_data['total_vaccinations']) ax1.set_xticks(x1) ax1.set_title('United States: Pfizer', size=25) ax1.set_xlabel('Date') ax1.set_ylabel(''Total vaccinations x100000000') ax2.plot(Germany_data['date'], Germany_data['total_vaccinations']) x2 = range(0,len(Germany_data['date']),25) ax2.plot(Germany_data['date']),Germany_data['total_vaccinations']) ax2.set_xticks(x2) ax2.set_title('Germany: Pfizer', size=25) ax2.set_xlabel('Date') ax2.set_ylabel(''Total vaccinations x100000000') plt.show() |
This concludes the explanation of data processing using Frovedis DataFrame.